About me
My name is Verena Kaynig-Fittkau and I am a Post-Doc fellow at Harvard School of Engineering and Applied Sciences, in Cambridge, MA in the GVI Lab of Hanspeter Pfister.
I recieved my PhD in computer science in 2011 from ETH Zurich, where I worked on Connectomics in the Pattern Analysis and Machine Learning Group headed by Prof. Joachim M. Buhmann. I also developed image processing approaches for electron microscopy images for the EMEZ (Electron Microscopy ETH Zurich).
I recieved my diploma in computer science in 2006 from the University of Hamburg. During undergraduate studies I worked for the EU Projects ViSiCAST and CogVIS. During that time I also worked as a parttime programmer for Beiersdorf R&D, developing tools for stitching and alignment of light and electron microscopy images.
Research interests:
- Biomedical image processing
- Structure preserving image registration
- Image segmentation
- Feature selection
Research
Goal of my PhD project is to develop image processing and machine learning methods and algorithms to detect and classify and reconstruct neuroanatomical structures like neurons, dendrites and synapses from transmission electron microscopy images. We work in close cooperation with the Institute of Neuroinformatics UniZH | ETH Zurich (INI) and electron microscopy ETH Zurich (EMEZ). In order to get a better understanding of learning and remembering in the brain, neuroanatomists build 3d reconstructions of brain tissue from electron microscopy images. Today most of this work involves time consuming manual image processing, which renders the process very tedious and time consuming. Automating the image processing pipeline will not only make this process faster, but enable the anatomist to look at much larger datasets, hopefully leading to new insights of the functional structure of the brain.
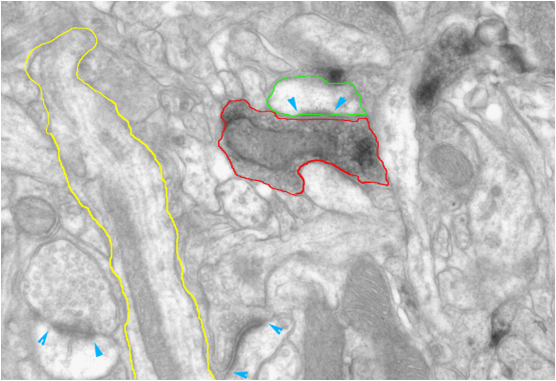
Figure 1: Example of a TEM image of neuronal tissue. Segmented structures are: dendrite (yellow), chemically labeled bouton (red) and a spine head (green). Blue arrows point to synapses. Image courtesy of Kevan Martin and Nuno Miguel Macarico A. da Costa (UniZH | ETH Zurich).
Image processing workflow
This diagram shows the image processing pipeline for building 3d reconstructions out of TEM images. First, single images are taken from serial sections. The electromagnetic lenses of the microscope induce a non linear transform leading to distorted images. We correct the images against these distortions. As the camera range is limited, multiple images are taken from the same section. These images need to be stitched together. The next step is the z-aligment of section images. Finally the structures of interest in the image stack need to be segmented and reconstructed in 3d.
Publications
- Kaynig, V., Fuchs, T., Buhmann, J. M., Geometrical Consistent 3D Tracing of Neuronal Processes in ssTEM Data , MICCAI, 2010
- Kaynig, V., Fischer, B., Müller, E., Buhmann, J. M., Fully Automatic Stitching and Distortion Correction of Transmission Electron Microscope Images , Journal of Structural Biology, 171(2), August 2010, 163-173. PDF
- Kaynig, V., Fuchs, T., Buhmann, J. M., Neuron Geometry Extraction by Perceptual Grouping in ssTEM Images, CVPR, 2010. PDF
- Kaynig, V., Fischer, B., Buhmann, J. M., Probabilistic Image Registration and Anomaly Detection by Nonlinear Warping, CVPR, 2008. PDF
- Kaynig, V., Fischer, B., Wepf, R. & Buhmann, J. M., Fully Automatic Registration of Electron Microscopy Images with High and Low Resolution, Microscopy and Microanalysis, 2007. PDF
Software
Lens distortion correction
The electromagnetic lenses of transmission electron microscopes induce a non linear transform to the images. We developed a calibration method to correct these distortions. The code is open source and available as a plugin for imagej. It comes as part of fiji and can be found under Plugins - Transform - Distortion Correction. A documentation how to use the plugin can be found here
